Billions of years ago, before there were beasts, bacteria or any living organism, there were RNAs. These molecules were probably swirling around with amino acids and other rudimentary biomolecules, merging and diverging, on an otherwise lifeless crucible of a planet.
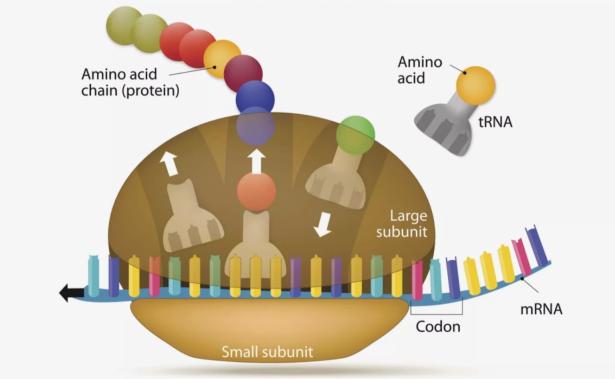
Then, somehow, something special emerged: a simple machine, a pocket made of RNAs, with the ability to place amino acids next to one another and maybe link them into chains. This was the macromolecule that would gradually evolve into the ribosome, the RNA–protein complex responsible for translating genetic information into proteins. Its birth — the details of which remain hypothetical — would have created a fundamental shift in this prebiotic, RNA-dominated world, providing a key ingredient to all life as we know it. Ada Yonath, a structural biologist at the Weizmann Institute of Science in Rehovot, Israel, and her team first conceptualized this ‘protoribosome’ idea nearly two decades ago, after she and others determined the structure of the modern ribosome, a feat that later secured Yonath a share of the 2009 Nobel Prize in Chemistry.
But to solidify the case for the hypothetical protoribosome, Yonath and her laboratory would have to build it.
It’s a project that other scientists have watched with interest. And the lab’s achievements in the past two years — creating a primitive RNA machine that can link two amino acids together1,2 — have created a ripple of excitement. A group in Japan, working separately and led by molecular biologist Koji Tamura at the Tokyo University of Science, has succeeded in creating a similar, functional protoribosome3. “It’s strongly supportive of Ada’s protoribosome idea,” says Paul Schimmel, a biochemist at Scripps Research in La Jolla, California. “I’ve always had an open mind; the open mind is becoming a more convinced mind.”
Although doubts and caveats remain, Yonath’s and Tamura’s work seems to recapitulate a milestone on the road from primordial organic molecules to the ribosome used by the last common ancestor of all living things. This was no simple task: In Yonath’s group, the project was passed from researcher to researcher and took more than 15 years to succeed. The work has now opened the door for origin-of-life scientists to fill in further details. And others are looking at the protoribosome, or something like it, as a tool to create new kinds of biomolecule.
“This should be a starting point of many more fields of research,” says Tanaya Bose, a postdoctoral researcher who has led the Yonath group’s efforts for the past several years.
Crystal inspiration
Scientists have been attempting to recreate some semblance of the chemical origin of biomolecules for decades; it was 70 years ago that Stanley Miller, a chemist at the University of Chicago in Illinois, sparked a gas mixture to create organic compounds4. Researchers including Carl Woese and Francis Crick suggested the ribosome could have started as a molecule made solely of RNA, an idea that was supported 40 years ago with the proof that RNAs can catalyse reactions5,6. This led to the ‘RNA world’ hypothesis, which describes a time, before cells or actual life, when RNAs replicated and catalysed reactions7. The RNA-world idea has come into doubt in the past decade; many scientists now suspect that a variety of biomolecules, including rudimentary proteins, lipids and metabolites, might have existed together with nucleic acids.
Yonath suspects there were plenty of RNAs on early Earth. “Most of them don’t exist any more because they were small and not useful.” The protoribosome, she says, was the one that lasted.
Yonath and her colleagues published high-resolution structures for the two protein–RNA subunits that make up a ribosome in the early 2000s8,9. These examples came from extremophile bacteria. And as ribosomal structures from other organisms were published, Ilana Agmon, a scientific adviser in Yonath’s group, noticed something that she found striking. Deep in the core of the large subunit was a semi-symmetrical segment. This region contained a pocket-like structure, made of ribosomal RNA, called the peptidyl transferase centre (PTC). During the translation of mRNA into protein, when two amino acids are placed in the PTC, it creates conditions for them to click together. And, although the specific nucleotide sequence of the structure varied between species, the shape was the same in every example, suggesting that it’s crucial to the ribosome’s ability to support life (see ‘The heart of the matter’).
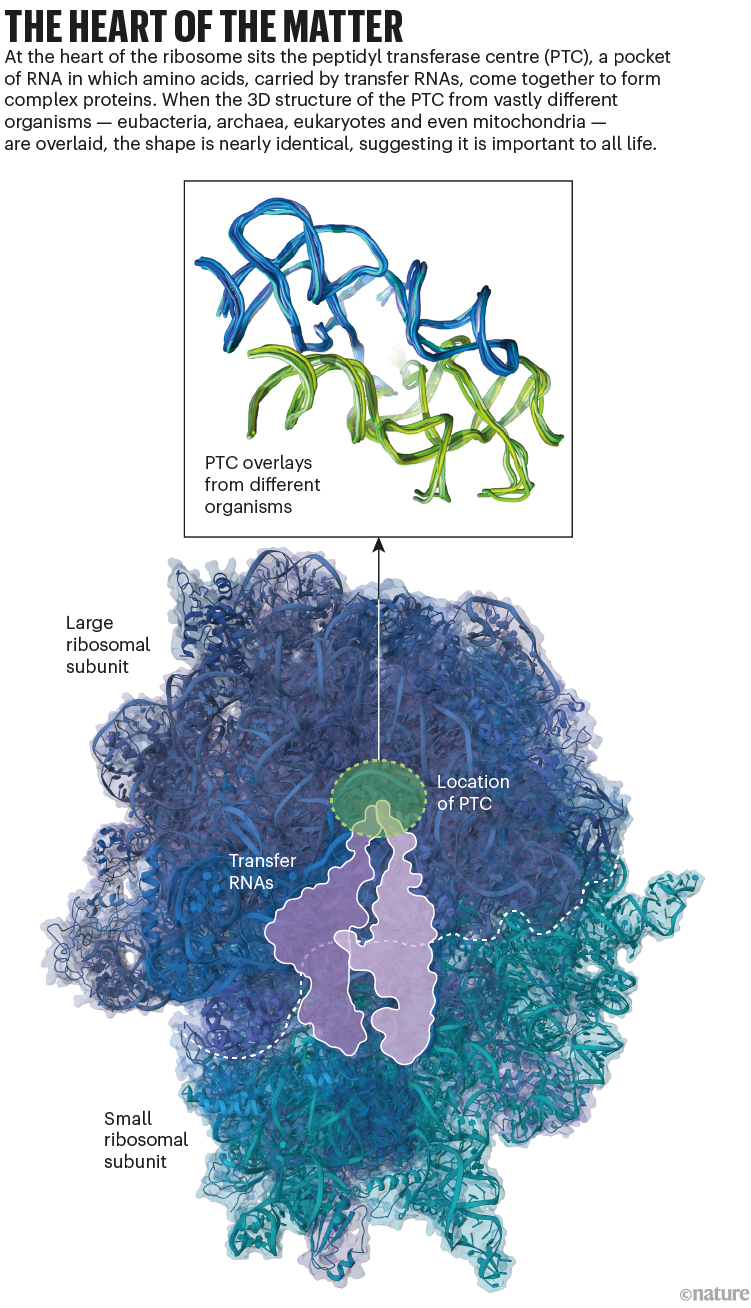
Source: refs 1 & 3/PDB.
In 2006, when Yonath started asking her team about the evolution of the ribosome, Agmon suggested looking closely at the semi-symmetrical region containing the PTC. “We speculated that this is the protoribosome, this is the part from which the ribosome evolved,” recalls Anat Bashan, a senior staff scientist who’s been involved in the project since the beginning. But, the region they were positing as the putative protoribosome is assembled from 178 ribonucleotides, which Yonath noted would be an awfully large structure to spring into being, fully formed, on primordial Earth. Based in part on the symmetry she’d observed, Agmon proposed a model requiring 2 similar, L-shaped RNAs of 60 and 61 nucleotides. The team thought this was a more reasonable size of molecule to arise on early Earth.
Fragments of that size are certainly plausible. Elisa Biondi, a biochemist at the Foundation for Applied Molecular Evolution in Alachua, Florida, and her colleagues managed to synthesize RNAs of about 100–300 bases long on materials called rock glasses, which would have formed owing to volcanic action or meteor impacts in a prebiotic world10.
But not everyone is convinced that Yonath’s and Agmon’s short fragments arose spontaneously. Joseph Moran, an organic chemist at the University of Strasbourg in France, praises Yonath’s accomplishments but doubts that the protoribosome just popped into existence. “It had to be derived from much simpler things,” he says.
Robert Root-Bernstein, a biologist at Michigan State University in East Lansing, has a theory for what those simpler things might have been: transfer RNAs (tRNAs). In the modern ribosome, amino acids enter the PTC attached to tRNAs that match the three-letter codes on the mRNA. Those codes determine which amino acid comes next in the protein. To Root-Bernstein and others, the PTC core looks a lot like four tRNAs joined together. And tRNAs, he notes, don’t just deliver amino acids to the ribosome; they are versatile molecules that can do all kinds of task, such as sensing nutrients and silencing genes. Perhaps they had some function before the protoribosome, then provided the building blocks for that structure.
Extraordinary evidence
Regardless of how the hypothetical protoribosome came to be, at the time when Agmon and Yonath first conceived it, there was no experimental evidence to show that it could have existed and worked as they thought it did. The protoribosome hypothesis assumes that one of those early RNA pockets could link amino acids together, and that it then evolved into the ribosome, which has that same function. Scientists who spoke with Nature say that’s a fair idea, but it’s not a given. “It’s plausible,” says Anton Petrov, an evolutionary biologist at the Georgia Institute of Technology in Atlanta, but he also thinks that early RNA machines might have had functions distinct from peptide synthesis, and that they then took on that role later as the protoribosome emerged.
“It was a very bold and imaginative suggestion from Yonath,” says John Sutherland, a chemist at the Medical Research Council Laboratory of Molecular Biology in Cambridge, UK.
Oldest-ever DNA shows mastodons roamed Greenland 2 million years ago
Bold theories require extraordinary evidence, and that’s what the lab set out to obtain. After developing the protoribosome hypothesis, Agmon moved to the Technion — Israel Institute of Technology in Haifa. Graduate student Chen Davidovich took the lead as the experiments commenced. The first step, Yonath says, was to produce the molecules to build this theoretical protoribosome.
Davidovich studied the RNA sequences of various modern ribosomes. The macromolecule contains dozens of ribonucleotides and accessory proteins, but not all of these are involved in the PTC’s shape or functions. He stripped out whatever seemed like it would be extraneous to the protoribosome, leaving just enough RNA to create that semi-symmetrical pocket2. Some of these RNAs were able to pair up into something similar to the PTC core that Agmon had imagined.
“This took a long time,” Yonath recalls — but the next step would take even longer. The second step was to show that these putative protoribosomes could take two amino acids and hook them together.
Davidovich attempted to show PTC activity in the same way that other researchers have measured the ability of the modern ribosome to link two amino acid analogues into a dipeptide. He labelled such analogues, linked to a handful of nucleotides to stand in for tRNA, with radioactivity. After mixing them with the protoribosome, he thought he would be able to sort the molecules by size and find longer, radio-labelled dipeptides. But he never saw a hint of dipeptide. “It almost killed my career,” recalls Davidovich. Fortunately, he had back-up projects related to another interest of Yonath’s lab, antibiotics that attack ribosomes. He graduated in 2010 and now leads a lab studying gene repression at Monash University in Melbourne, Australia.
Next up was graduate student Miri Krupkin, who was drawn in by the symmetry of the PTC and its ability to build proteins quicker than any human chemist has managed. Using Davidovich’s constructs and others that Krupkin designed herself, she too attempted to detect the radioactive dipeptide product on the basis of on its size. She also tried labelling the substrates with fluorescence, instead of radioactivity. Still, nothing.
Krupkin began to wonder if the protoribosome even made peptides when it first emerged. She tested other potential chemistries, such as adding or removing phosphate groups from other molecules, and still came up dry.
Origin of life theory involving RNA–protein hybrid gets new support
“I kept going, because it’s such an interesting question,” says Krupkin. But similar to Davidovich, she had to rely on another project, also on antibiotics, to graduate in 2016. She’s currently investigating HIV’s RNA structure as a postdoc research fellow at Stanford University in California.
Bose, who joined the lab as a postdoc in 2016, would take the project to the finish line. She had trained as a chemist, and that gave her a fresh perspective. She knew that if the primitive protoribosome worked at all, it would probably be inefficient, yielding a minute amount of dipeptide. Rather than separating the reaction products by size, Bose turned to mass spectrometry, which had advanced to become the most sensitive method by this time. It still wasn’t easy — she performed many reactions and control experiments, and tried two forms of mass spectrometry — but at last, she saw a peak representing the expected dipeptide1,2.
“The amount of product was tiny,” says Bose. She suspects Davidovich and Krupkin might have been making it all along, but there wasn’t enough to detect it using their methods.
Even that tiny amount was “a bit of a feat” to produce, says Sutherland. “They’ve made progress on a really difficult problem.”
Next steps
By the time Krupkin was trying to measure a dipeptide product, the Yonath group wasn’t the only one on the trail of the minimal peptide-bonding machine. Tamura, in Japan, was also inspired by the semi-symmetrical core pocket. He recruited a group including master’s student Mai Kawabata and undergraduate Kentaro Kawashima to make their own protoribosome-like structure.
Their PTC facsimile, made from two 74-nucleotide RNAs, was similar to Yonath’s, but to stand in for modern tRNAs, they used structures called minihelices. These are about half the size of a modern tRNA, and were therefore considerably larger than the tRNA replacements Yonath used. Modern tRNAs are thought to have evolved from minihelices.
Tamura’s team eventually achieved its own success, also detecting the dipeptide with mass spectrometry3. “Mai and Kentaro were patient and responsive, and finally, we won a big win,” says Tamura.
Schimmel, Tamura’s former postdoc adviser, says that the minihelices were a key addition. “He carried it to the next level, so to speak, in that he constructed what is believed by many evolutionists to be the primordial tRNA,” says Schimmel. Some scientists, he notes, even suspect that minihelices evolved the ability to self-replicate, another key step on the way to living organisms.
The shifting sands of ‘gain-of-function’ research
Tamura cautions that neither lab’s construct works exactly like the modern PTC does. “All of our results are too simple,” he says. “We still have a long way to go before we really understand the evolution of the PTC and the ribosome.”
For Bashan, it’s close enough. “We think this is how the first proteins came about,” she says.
But as other scientists note, there were probably other ways for polypeptides to emerge on early Earth. Linking up amino acids is “dead easy”, says Sutherland. For example, some scientists have proposed that amino acids plus α-hydroxy acids (a group that includes, for example, lactic acid and citric acid), could have formed into polypeptides during cycles of cool, wet and hot, dry conditions on early Earth — with no RNA required11. And last year, another team used RNA bases, although not the A, C, G, and U ones used in modern RNA codes, to put peptides together. The researchers proposed this could have happened, without the ribosome, in the RNA world12. The catch, says Sutherland: “it bore no resemblance to the way that nature now makes peptides from RNA”.
What is special about the protoribosome work, say Sutherland and Yonath, is that it is possible to imagine how this primitive core, over millennia, might have accumulated extra pieces of RNA and protein to create the modern ribosome.
Petrov and his colleagues have predicted exactly that kind of timeline by working backwards from today’s ribosome. They analysed ribosomes for atomic-level ‘fingerprints’ that were left in the 3D ribosome structure when new branches of RNA were added13. This led to a model that showed early structures matching those of Yonath’s protoribosome, with extra pieces of RNA slowly incorporated over time (see ‘Molecular evolution’).
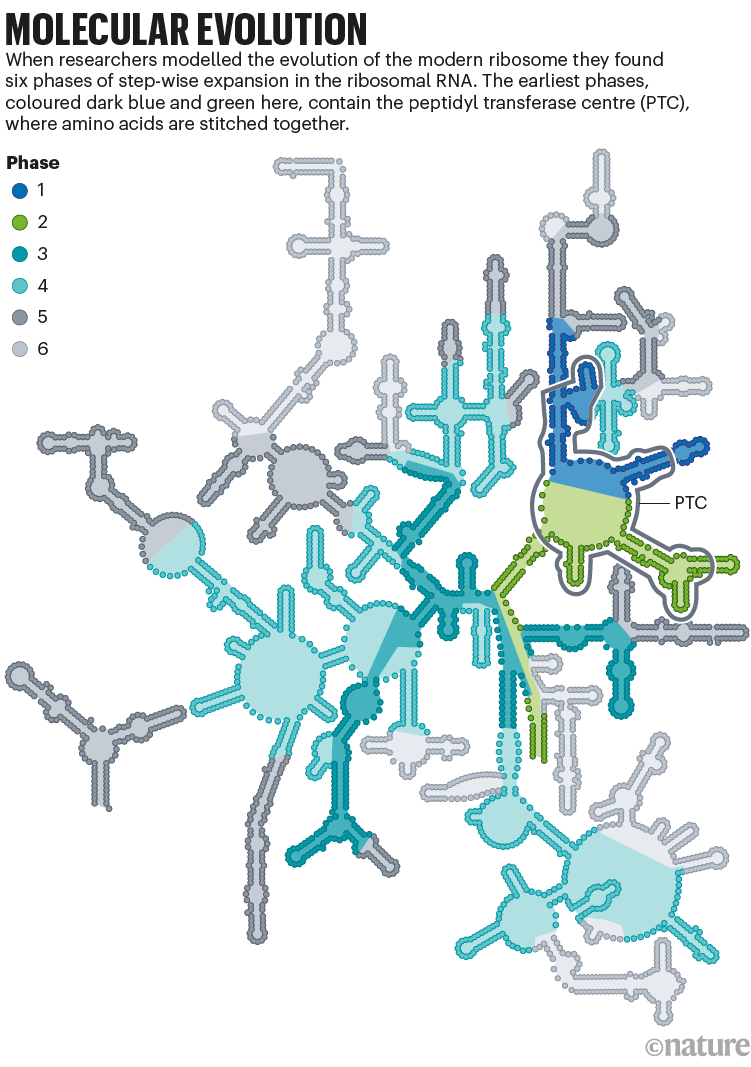
Source: ref. 13.
One obvious next step for Yonath’s team is to try to make a diverse set of peptides that are longer than two amino acids. Bose is working on this, but it will require different starting materials.
There is one group that claims to have synthesized a longer peptide from a protoribosome-like structure. Yuhong Wang, a biophysicist at the University of Houston in Texas, and her colleague reported mass spectrometry results from experiments with a larger version of the PTC that indicate the production of a chain of nine lysines14. Wang thinks that there might be lysine polymers of other lengths as well. “I think we have strong evidence,” she says, but she admits that the controls are incomplete, and she cannot explain why strings of nine amino acids would be the most prominent product.
Wang and others are interested in using such stripped-down ribosomes to manufacture new kinds of biomolecule, not limited to the usual 20 amino acids or even to amino acids at all, that could have uses in medicine or industry. For example, they might build molecules out of amino acids that are right-handed in structure, unlike the left-handed ones seen in life on Earth. This kind of macromolecule synthesis could be cheaper and more environmentally friendly than other means, Wang suggests.
Meanwhile, there’s plenty of research left to do for origin-of-life studies. Scientists need to work out how RNAs gained the ability to self-replicate. And they need to discover how an early ribosome would have managed to create specific peptides encoded by primitive mRNAs. Those processes, plus the ability to synthesize peptides, would provide the raw materials for evolution.
There’s one more factor, Sutherland says: the early peptides made by the protoribosome must have been somehow useful, or there would be no evolutionary advantage to the machine’s continued existence. He suggests a couple of speculative functions: perhaps the peptides sequestered metal ions that would otherwise destroy RNAs. Or, they might have helped to form early biomolecular compartments to concentrate RNA and peptides together.
“When you get something that evolution can act on,” says Sutherland, “the rest is history.”
Nature 615, 22-25 (2023)
doi: https://doi.org/10.1038/d41586-023-00574-4
Amber Dance is a freelance science journalist in the Los Angeles area.
References
-
Bose, T., Fridkin, G., Bashan, A. & Yonath, A. Israel J. Chem. 61, 863–872 (2021).
-
Bose, T. et al. Nucleic Acids Res. 50, 1815–1828 (2022).
-
Kawabata, M. et al. Life 12, 573 (2022).
-
Miller, S. L. Science 117, 528–529 (1953).
-
Kruger, K. et al. Cell 31, 147–157 (1982).
-
Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. Cell 35, 849–857 (1983).
-
Gilbert, W. Nature 319, 618 (1986).
-
Schluenzen, F. et al. Cell 102, 615–623 (2000).
-
Harms, J. et al. Cell 107, 679–688 (2001).
-
Jerome, C. A., Kim, H.-J., Mojzsis, S. J., Benner, S. A. & Biondi, E. Astrobiology 22, 629–636 (2022).
-
Forsythe, J. G. et al. Angew. Chem. Int. Edn Engl. 54, 9871–9875 (2015).
-
Müller, F. et al. Nature 605, 279–284 (2022).
-
Petrov, A. S. et al. Proc. Natl Acad. Sci. USA 111,10251–10256 (2014).
-
Xu, D. & Wang, Y. Biochem. Biophys. Res. Commun. 544, 81–85 (2021).



Spread the word