Early in his career, the decorated biologist Richard Lenski thought he might be forced to evolve. After his postdoctoral research grant was canceled, Lenski began to look tentatively at other options. With one child and a second on the way, Lenski attended a seminar about using specific types of data in an actuarial context — the same type of data he had worked with as a graduate student. Lenski collected a business card from the speaker, thinking he might be able to make use of his background in a new career.
“But then, as it sometimes does — and I was very lucky — the tide turned,” Lenski told Quanta Magazine in his high-rise office at Michigan State University. “We got the grant renewed, and soon thereafter, I started getting faculty offers.”
Lenski, a professor of microbial ecology at Michigan State, is best known for his work on what’s known as the long-term evolution experiment. The project, started in 1988, examines evolution in action. He and his lab members have been growing 12 populations of E. coli continuously for over 65,000 generations, tracking the development and mutations of the 12 separate strains.
The results have garnered attention and accolades — including a MacArthur “genius” grant, which Lenski received in 1996 — both for the enormity of the undertaking and for the intriguing findings the study has yielded. Most notably, in 2003, Lenski and his collaborators realized that one strain of E. coli had evolved the ability to use citrate as an energy source, something no previous population of E. coli was able to do.
Lenski is also interested in digital organisms, computer programs that have been designed to mimic the process of evolution. He was instrumental in the push to open the Beacon Center at Michigan State, which gives computer scientists and evolutionary biologists the opportunity to forge unique collaborations.
Quanta Magazine met with Lenski in his office to talk about his own evolving interests in the field of evolutionary biology — and about the time he almost pulled the plug on the long-term experiment. An edited and condensed version of the conversation follows.
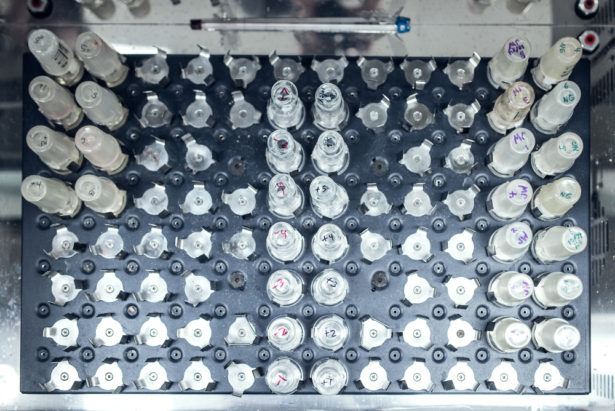
Logan Zillmer for Quanta Magazine. Vials containing the E. coli strains that make up the long-term evolution experiment.
QUANTA MAGAZINE: What sort of questions have been driving forces in your career?
RICHARD LENSKI: One question that has always intrigued me is about the reproducibility or repeatability of evolution. Stephen Jay Gould, the paleontologist and historian of science, posed this question: If we could rewind the tape of life on Earth, how similar or dissimilar would it be if we watched the whole process play out again? The long-term experiment that we do has allowed us to gather a lot of data about this question.
So is evolution repeatable?
Yes and no! I sometimes tell people it’s been a fascinating motivating question, but on one level, it is a terrible question, and one you would never tell a graduate student to go after. That’s because it is very open-ended, and it does not have a very clear-cut answer.
From the long-term experiment, we’ve seen some really beautiful examples of things that are remarkably reproducible, and on the other hand some other crazy things where one population goes off and does things that are entirely different from the other 11 populations in the experiment.
How did you first come up with the idea for the long-term experiment?
I had been working already for several years on experimental evolution with bacteria, as well as viruses that infect bacteria. Those were fascinating, but everything became so complicated so quickly that I said, “Let’s reduce evolution down to its bare bones.” In particular, I wanted to go after this question of reproducibility or repeatability of evolution. And if I wanted to be able to look at the reproducibility of evolution, I wanted a system that was very simple. When I started the long-term experiment, my original goal was that I would call it the long-term experiment when I got to 2,000 generations.
How long did that take you?
The actual running of the experiment was about 10 or 11 months, but by the time we had collected data, wrote it up, and got the paper published, it was more like two and a half years or so. By then the experiment had already passed 5,000 generations, and I realized we should keep it going.
Did you anticipate the experiment going on for as long as it has?
No. No, I didn’t. There was a five-year period, maybe from the late ’90s into the early 2000s, where I thought about possibly stopping the experiment. This was for a couple of different reasons. One was that I was getting hooked on this other way of studying evolution, which involved looking at evolution in self-replicating computer programs, which was absolutely fascinating. Suddenly I saw this even shinier way of studying evolution, where it could go even more generations and do even more, seemingly neater, experiments.
How have your views on studying evolution via these digital organisms changed over time?
I had this sort of “puppy love” when I first learned about it. At first, it was just so extraordinarily interesting and exciting to be able to watch self-replicating programs, to be able to change their environments, and to watch evolution happen.
One of the really exciting things about digital evolution is that it shows that we think of evolution as being about stuff with blood and guts and DNA and RNA and proteins. But the idea of evolution really comes down to some very basic ideas of heredity, replication and competition. The philosopher of science Daniel Dennett has emphasized that we see evolution as this instantiation, this form of biological life, but the principles of it are much more general than that.
I would say that my latest directions of research have been primarily by way of talking with super-smart colleagues and serving on committees of graduate students who are using these systems. I’m less involved in designing experiments or formulating specific hypotheses, because that field has been moving extremely quickly. I feel I was very lucky to pick off some of the low-hanging fruit, but now I feel like I’m in there as a biologist, maybe criticizing hypotheses, suggesting controls that might be done in some experiments.
So your interest in digital organisms was one reason you considered shutting down the long-term experiment. What was the other?
At that point, the other thing that was a little frustrating about the long-term lines was that the rate at which the bacteria were changing was slowing down. The way I thought about it, it was almost as though evolution had stopped. I thought that this was just too simple an environment, and there wasn’t that much more for them to do.
So those two different things made me think about stopping the experiment. And I spoke to a few colleagues and they basically told me: You can’t do that. You shouldn’t do that. I talked with my wife, Madeleine, by the way, when I was getting very interested in these digital organisms — we were actually on sabbatical in France at that time — and I said, “Maybe I should call home and shut down the lab.” And she said, “I don’t think you should do that.”
Why did your wife and your colleagues have that reaction?
The experiment had already been quite profitable in a scientific sense, providing very rich data about the dynamics of evolutionary change. It was more or less unique in the timescales it was probing. So I think it was very good advice they gave me. I don’t know whether I could have ever quite pulled the plug myself. I certainly was a bit frustrated and thinking about it — but anyhow, people said no!
Logan Zillmer for Quanta Magazine. Video: Lenski discusses how he has been surprised by evolution.
Did you get past the plateau where you said you felt like the organisms weren’t evolving that much?
That actually has been one of the really cool findings from the experiment. When I started the long-term experiment, I thought that the bacteria would quickly reach some sort of limit to their growth. It was only a few years ago that we began to realize that the bacteria would always be able to beat anything we had inferred in the past about what their hard limit might be. I realized that we’re just fundamentally not thinking about this the right way. Even in the simplest environment, there’s always the potential for organisms to do any step in their metabolism, or any step in their biochemistry, a little bit better. And natural selection, although it won’t get it right on any given step, will over the long term always be favoring these subtle improvements.
One line of bacteria evolved the ability to use citrate as a food source. Did that happen before or after you were thinking of shutting down the experiment?
That was one of the things that made me realize we wouldn’t shut down the experiment. In 2003, one lineage evolved the ability to use citrate. That became a game changer: realizing that even in this super simple environment, there were some major things for the bacteria to evolve and figure out.
I like to say that the bacteria would eat dinner every night without realizing there was this nice, lemony dessert right around the corner. And so far, even after 65,000 generations, only one of the 12 populations has figured out how to consume that citrate.
You also mentioned that certain populations within your experiment have developed mutations at a greater rate. What does that look like?
After over 60,000 generations, six out of the 12 populations have evolved to be hypermutable. They’ve evolved changes in their DNA repair and DNA metabolic processes that causes them to have new mutations somewhere on the order of 100 times the rate at which the ancestor [at the start of the experiment] did.

Logan Zillmer for Quanta Magazine. Lenski's laboratory at Michigan State University.
It’s a very interesting process, because it’s both good and bad from the bacteria’s perspective. It’s bad because most mutations are harmful or at best neutral. Only the rare nugget in the mine is a beneficial mutation. The bacteria that have the higher mutation rate are a little bit more likely to discover one of those nuggets. But on the other hand, they’re also more likely to produce children and grandchildren with deleterious mutations.
Was the line that was able to consume citrate part of the group that had evolved to be hypermutable?
That’s a great question. The line that evolved the ability to use citrate did not have an elevated mutation rate. Interestingly, it became one of the ones with a higher mutation rate, but only after it evolved the ability to use citrate. It’s consistent with the benefit of the higher mutation rate — the additional capacity for exploration. The bacteria were actually quite poor at using citrate to begin with, so there were a lot of opportunities after they evolved the ability to use citrate to refine that ability.
How does the long-term experiment help you understand the evolution of life on a larger scale?
For me, one of the lessons of the long-term experiment has been how rich and interesting life can be, even in the dullest, simplest environment. The fact that evolution can generate this diversity, and discover doors left slightly ajar that it can push through, speaks to the awesome inventiveness of evolution. And if it can be so inventive and creative on this minuscule spatial and temporal scale, and in such a dull environment, it just creates more awe in me, when I think of how much more remarkable it is out in nature.
What most surprised you about this project?
That it’s still going on after all these years. One of my goals in life is to make sure that the experiment continues. I would like to raise an endowment to keep the experiment going on in perpetuity.
What’s your hope for the long-term experiment in the future?
My hope for the project is that it will yield many more surprises. For instance, two lineages have coexisted for 60,000 generations in one of the populations, where one of them is feeding off of the product that the other one is generating. I think it’s fascinating to wonder if, at some point, that might turn into something more like a predator-prey interaction. It’s certainly not outside the realm of possibilities. Whether it would ever happen, I don’t know.
It has also been a tremendous joy to work with students, postdocs and collaborators, and to see them grow and develop. That’s really the biggest joy for me of being a scientist. I like to tell people that I’m a bigamist. I have two families: I have my lab family and my biological family, and they both are incredibly wonderful.
Quanta Magazine's mission is to enhance public understanding of research developments in mathematics and the physical and life sciences. Quanta articles do not necessarily represent the views of the Simons Foundation. Learn more



Spread the word