For someone who’s not a Sherlock superfan, cognitive neuroscientist Janice Chen knows the BBC’s hit detective drama better than most. With the help of a brain scanner, she spies on what happens inside viewers’ heads when they watch the first episode of the series and then describe the plot.
Chen, a researcher at Johns Hopkins University in Baltimore, Maryland, has heard all sorts of variations on an early scene, when a woman flirts with the famously aloof detective in a morgue. Some people find Sherlock Holmes rude while others think he is oblivious to the woman’s nervous advances. But Chen and her colleagues found something odd when they scanned viewers’ brains: as different people retold their own versions of the same scene, their brains produced remarkably similar patterns of activity1.
Chen is among a growing number of researchers using brain imaging to identify the activity patterns involved in creating and recalling a specific memory. Powerful technological innovations in human and animal neuroscience in the past decade are enabling researchers to uncover fundamental rules about how individual memories form, organize and interact with each other. Using techniques for labelling active neurons, for example, teams have located circuits associated with the memory of a painful stimulus in rodents and successfully reactivated those pathways to trigger the memory. And in humans, studies have identified the signatures of particular recollections, which reveal some of the ways that the brain organizes and links memories to aid recollection. Such findings could one day help to reveal why memories fail in old age or disease, or how false memories creep into eyewitness testimony. These insights might also lead to strategies for improved learning and memory.
The work represents a dramatic departure from previous memory research, which identified more general locations and mechanisms. “The results from the rodents and humans are now really coming together,” says neuroscientist Sheena Josselyn at the Hospital for Sick Children in Toronto, Canada. “I can’t imagine wanting to look at anything else.”
In search of the engram
The physical trace of a single memory — also called an engram — has long evaded capture. US psychologist Karl Lashley was one of the first to pursue it and devoted much of his career to the quest. Beginning around 1916, he trained rats to run through a simple maze, and then destroyed a chunk of cortex, the brain’s outer surface. Then he put them in the maze again. Often the damaged brain tissue made little difference. Year after year, the physical location of the rats’ memories remained elusive. Summing up his ambitious mission in 1950, Lashley wrote2: “I sometimes feel, in reviewing the evidence on the localization of the memory trace, that the necessary conclusion is that learning is just not possible.”
Memory, it turns out, is a highly distributed process, not relegated to any one region of the brain. And different types of memory involve different sets of areas. Many structures that are important for memory encoding and retrieval, such as the hippocampus, lie outside the cortex — and Lashley largely missed them. Most neuroscientists now believe that a given experience causes a subset of cells across these regions to fire, change their gene expression, form new connections, and alter the strength of existing ones — changes that collectively store a memory. Recollection, according to current theories, occurs when these neurons fire again and replay the activity patterns associated with past experience.
Scientists have worked out some basic principles of this broad framework. But testing higher-level theories about how groups of neurons store and retrieve specific bits of information is still challenging. Only in the past decade have new techniques for labelling, activating and silencing specific neurons in animals allowed researchers to pinpoint which neurons make up a single memory (see ‘Manipulating memory’).
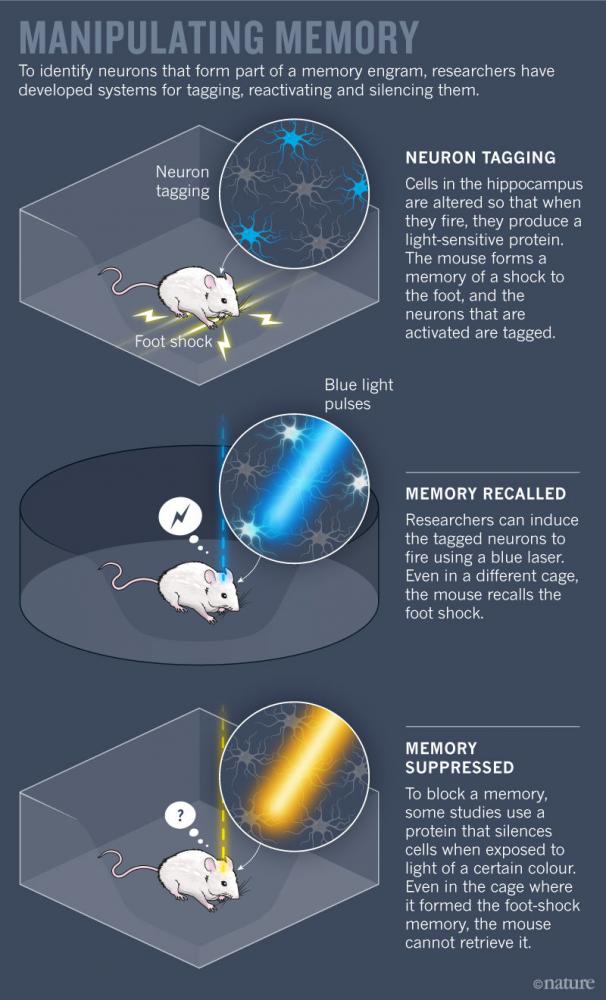
Josselyn helped lead this wave of research with some of the earliest studies to capture engram neurons in mice3. In 2009, she and her team boosted the level of a key memory protein called CREB in some cells in the amygdala (an area involved in processing fear), and showed that those neurons were especially likely to fire when mice learnt, and later recalled, a fearful association between an auditory tone and foot shocks. The researchers reasoned that if these CREB-boosted cells were an essential part of the fear engram, then eliminating them would erase the memory associated with the tone and remove the animals’ fear of it. So the team used a toxin to kill the neurons with increased CREB levels, and the animals permanently forgot their fear.
A few months later, Alcino Silva’s group at the University of California, Los Angeles, achieved similar results, suppressing fear memories in mice by biochemically inhibiting CREB-overproducing neurons4. In the process, they also discovered that at any given moment, cells with more CREB are more electrically excitable than their neighbours, which could explain their readiness to record incoming experiences. “In parallel, our labs discovered something completely new — that there are specific rules by which cells become part of the engram,” says Silva.
But these types of memory-suppression study sketch out only half of the engram. To prove beyond a doubt that scientists were in fact looking at engrams, they had to produce memories on demand, too. In 2012, Susumu Tonegawa’s group at the Massachusetts Institute of Technology in Cambridge reported creating a system that could do just that.
By genetically manipulating brain cells in mice, the researchers could tag firing neurons with a light-sensitive protein. They targeted neurons in the hippocampus, an essential region for memory processing. With the tagging system switched on, the scientists gave the animals a series of foot shocks. Neurons that responded to the shocks churned out the light-responsive protein, allowing researchers to single out cells that constitute the memory. They could then trigger these neurons to fire using laser light, reviving the unpleasant memory for the mice5. In a follow-up study, Tonegawa’s team placed mice in a new cage and delivered foot shocks, while at the same time re-activating neurons that formed the engram of a ‘safe’ cage. When the mice were returned to the safe cage, they froze in fear, showing that the fearful memory was incorrectly associated with a safe place6. Work from other groups has shown that a similar technique can be used to tag and then block a given memory7,8.
This collection of work from multiple groups has built a strong case that the physiological trace of a memory — or at least key components of this trace — can be pinned down to specific neurons, says Silva. Still, neurons in one part of the hippocampus or the amygdala are only a tiny part of a fearful foot-shock engram, which involves sights, smells, sounds and countless other sensations. “It’s probably in 10–30 different brain regions — that’s just a wild guess,” says Silva.
A broader brush
Advances in brain-imaging technology in humans are giving researchers the ability to zoom out and look at the brain-wide activity that makes up an engram. The most widely used technique, functional magnetic resonance imaging (fMRI), cannot resolve single neurons, but instead shows blobs of activity across different brain areas. Conventionally, fMRI has been used to pick out regions that respond most strongly to various tasks. But in recent years, powerful analyses have revealed the distinctive patterns, or signatures, of brain-wide activity that appear when people recall particular experiences. “It’s one of the most important revolutions in cognitive neuroscience,” says Michael Kahana, a neuroscientist at the University of Pennsylvania in Philadelphia.
The development of a technique called multi-voxel pattern analysis (MVPA) has catalysed this revolution. Sometimes called brain decoding, the statistical method typically feeds fMRI data into a computer algorithm that automatically learns the neural patterns associated with specific thoughts or experiences. As a graduate student in 2005, Sean Polyn — now a neuroscientist at Vanderbilt University in Nashville, Tennessee — helped lead a seminal study applying MVPA to human memory for the first time9. In his experiment, volunteers studied pictures of famous people, locations and common objects. Using fMRI data collected during this period, the researchers trained a computer program to identify activity patterns associated with studying each of these categories.
Later, as subjects lay in the scanner and listed all the items that they could remember, the category-specific neural signatures re-appeared a few seconds before each response. Before naming a celebrity, for instance, the ‘celebrity-like’ activity pattern emerged, including activation of an area of the cortex that processes faces. It was some of the first direct evidence that when people retrieve a specific memory, their brain revisits the state it was in when it encoded that information. “It was a very important paper,” says Chen. “I definitely consider my own work a direct descendant.”
Chen and others have since refined their techniques to decode memories with increasing precision. In the case of Chen’s Sherlockstudies, her group found that patterns of brain activity across 50 scenes of the opening episode could be clearly distinguished from one another. These patterns were remarkably specific, at times telling apart scenes that did or didn’t include Sherlock, and those that occurred indoors or outdoors.
Near the hippocampus and in several high-level processing centres such as the posterior medial cortex, the researchers saw the same scene-viewing patterns unfold as each person later recounted the episode — even if people described specific scenes differently1. They even observed similar brain activity in people who had never seen the show but had heard others’ accounts of it10.
“It was a surprise that we see that same fingerprint when different people are remembering the same scene, describing it in their own words, remembering it in whatever way they want to remember,” says Chen. The results suggest that brains — even in higher-order regions that process memory, concepts and complex cognition — may be organized more similarly across people than expected.
Melding memories
As new techniques provide a glimpse of the engram, researchers can begin studying not only how individual memories form, but how memories interact with each other and change over time.
At New York University, neuroscientist Lila Davachi is using MVPA to study how the brain sorts memories that share overlapping content. In a 2017 study with Alexa Tompary, then a graduate student in her lab, Davachi showed volunteers pictures of 128 objects, each paired with one of four scenes — a beach scene appeared with a mug, for example, and then a keyboard; a cityscape was paired with an umbrella, and so on. Each object appeared with only one scene, but many different objects appeared with the same scene11. At first, when the volunteers matched the objects to their corresponding scenes, each object elicited a different brain-activation pattern. But one week later, neural patterns during this recall task had become more similar for objects paired with the same scene. The brain had reorganized memories according to their shared scene information. “That clustering could represent the beginnings of learning the ‘gist’ of information,” says Davachi.
Clustering related memories could also help people use prior knowledge to learn new things, according to research by neuroscientist Alison Preston at the University of Texas at Austin. In a 2012 study, Preston’s group found that when some people view one pair of images (such as a basketball and a horse), and later see another pair (such as a horse and a lake) that shares a common item, their brains reactivate the pattern associated with the first pair12. This reactivation appears to bind together those related image pairs; people that showed this effect during learning were better at recognizing a connection later — implied, but never seen — between the two pictures that did not appear together (in this case, the basketball and the lake). “The brain is making connections, representing information and knowledge that is beyond our direct observation,” explains Preston. This process could help with a number of everyday activities, such as navigating an unfamiliar environment by inferring spatial relationships between a few known landmarks. Being able to connect related bits of information to form new ideas could also be important for creativity, or imagining future scenarios.
In a follow-up study, Preston has started to probe the mechanism behind memory linking, and has found that related memories can merge into a single representation, especially if the memories are acquired in close succession13. In a remarkable convergence, Silva’s work has also found that mice tend to link two memories formed closely in time. In 2016, his group observed that when mice learnt to fear foot shocks in one cage, they also began expressing fear towards a harmless cage they had visited a few hours earlier14. The researchers showed that neurons encoding one memory remained more excitable for at least five hours after learning, creating a window in which a partially overlapping engram might form. Indeed, when they labelled active neurons, Silva’s team found that many cells participated in both cage memories.
These findings suggest some of the neurobiological mechanisms that link individual memories into more general ideas about the world. “Our memory is not just pockets and islands of information,” says Josselyn. “We actually build concepts, and we link things together that have common threads between them.” The cost of this flexibility, however, could be the formation of false or faulty memories: Silva’s mice became scared of a harmless cage because their memory of it was formed so close in time to a fearful memory of a different cage. Extrapolating single experiences into abstract concepts and new ideas risks losing some detail of the individual memories. And as people retrieve individual memories, these might become linked or muddled. “Memory is not a stable phenomenon,” says Preston.
Researchers now want to explore how specific recollections evolve with time, and how they might be remodelled, distorted or even recreated when they are retrieved. And with the ability to identify and manipulate individual engram neurons in animals, scientists hope to bolster their theories about how cells store and serve up information — theories that have been difficult to test. “These theories are old and really intuitive, but we really didn’t know the mechanisms behind them,” says Preston. In particular, by pinpointing individual neurons that are essential for given memories, scientists can study in greater detail the cellular processes by which key neurons acquire, retrieve and lose information. “We’re sort of in a golden age right now,” says Josselyn. “We have all this technology to ask some very old questions.”
doi: 10.1038/d41586-018-00107-4
Sign up for the daily Nature Briefing email newsletter
References
-
Chen, J. et al. Nature Neurosci. 20, 115–125 (2016). PubMed Article CAS
-
Lashley, K. S. Soc. Exp. Biol. Symp. 4, 454–482 (1950). PubMed
-
Han, J. H. et al. Science 323, 1492–1496 (2009). PubMed Article CAS
-
Zhou, Y. et al. Nature Neurosci. 12, 1438–1443 (2009). PubMed Article CAS
-
Liu, X. et al. Nature 484, 381–385 (2012). PubMed Article CAS
-
Ramirez S. et al. Science 341, 387–391 (2013). PubMed Article CAS
-
Denny, C. A. et al. Neuron 83, 189–201 (2014). PubMed Article CAS
-
Tanaka, K. Z. et al. Neuron 84, 347–354 (2014). PubMed Article CAS
-
Polyn, S. M., Natu, V. S., Cohen, J. D. & Norman, K. A. Science310, 1963–1966 (2005). Article
-
Zadbood, A., Chen, J., Leong, Y. C., Norman, K. A. & Hasson, U. Cereb. Cortex 27, 4988–5000 (2017). Article CAS
-
Tompary, A. & Davachi, L. Neuron 96, 228–241 (2017). Article
-
Zeithamova, D., Dominick, A. L. & Preston, A. R. Neuron 75, 168–179 (2012). Article CAS
-
Zeithamova, D. & Preston, A. R. J. Cogn. Neurosci. 29, 1311–1323 (2017). PubMed Article
-
Cai, D. J. et al. Nature 534, 115–118 (2016). PubMed Article CAS


Spread the word